
NAStructural
Database
Structural database to facilitate computational studies of molecular modeling and recognition of proteins with special focus on antibody-antigen interactions.
Introduction
NAStructural DB is a specialized dataset designed to support predictive modeling in antibody research and wider in structural biology. It provides processed structures of antibodies, nanobodies, and proteins, along with their molecular contact information and essential annotations, including surface accessibility, secondary structure, and antibody region data. The dataset addresses key challenges such as sequence redundancy removal, inter- and intra-molecular contact mapping, and the need for a nanobody-specific subset. By streamlining data preparation, NAStructural DB aims to accelerate research in antibody-antigen interactions and therapeutic biologics development.
8 Deduplicated datasets
(4 complexes and 4 single chains) from 3 types of proteins(antibodies, nanobodies, non-antibody proteins)
antibody-antigen-interface - 1172 (from 1136 PDBs)
nanobody-antigen-interface - 487 (from 451 PDBs)
protein-protein-interface - 5158 (from 4453 PDBs)
heavy-light-interface - 2330 (from 2294 PDBs)
single-chain-heavy - 2164 (from 2135 PDBs)
single-chain-light - 1201 (from 1192 PDBs)
single-chain-nanobody - 602 (from 567 PDBs)
single-chain-non-ab - 23699 (from 22348 PDBs)
8 Full datasets
(4 complexes and 4 single chains) from 3 types of proteins(antibodies, nanobodies, non-antibody proteins)
full-antibody-antigen-interface - 2711 (from 1607 PDBs)
full-nanobody-antigen-interface - 1258 (from 620 PDBs)
full-protein-protein-interface - 47563 (from 16858 PDBs)
full-heavy-light-interface - 6217 (from 3758 PDBs)
full-single-chain-heavy - 6273 (from 3758 PDBs)
full-single-chain-light - 6654 (from 3928 PDBs)
full-single-chain-nanobody - 1825 (from 861 PDBs)
full-single-chain-non-ab - 422761 (from 170070 PDBs)
Pre-filtered
selecting only X-ray Crystallography with at least 3Å resolution
Annotated with
IMGT numbering
Secondary structure classification
Interaction characteristics
Surface exposure analysis
Molecular and chain associations
Molecular contact mapping
Accessing the Database
NAStructural DB is freely available for non-commercial organizations for non-commercial research. Commercial inquiries are welcome via contact us.
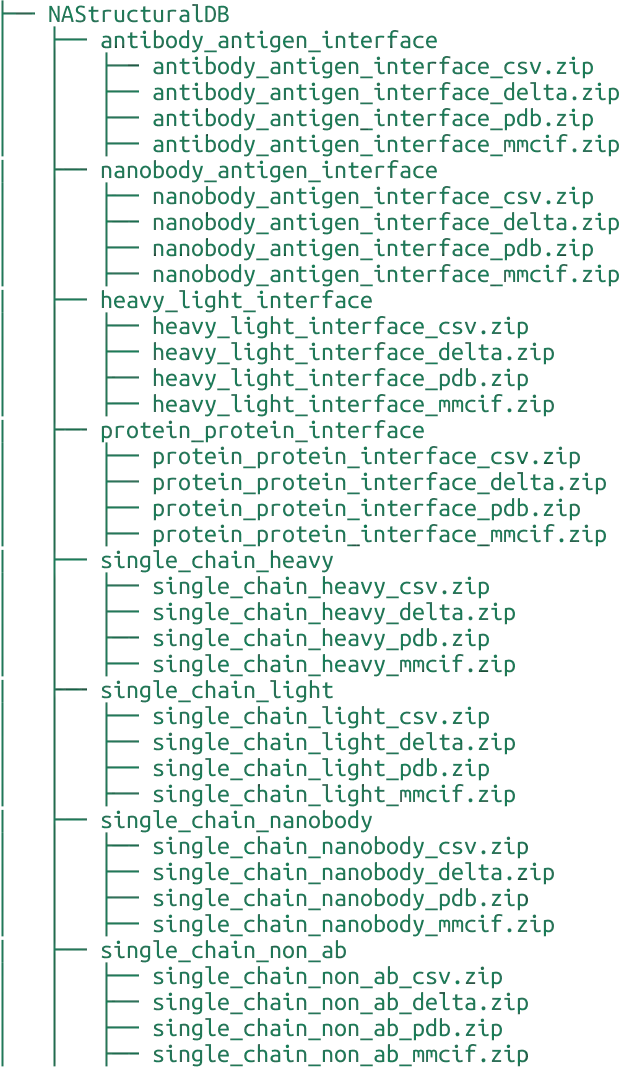
Directory structure
NAStructural DB is accessible using Google Drive with the following directory structure. It allows you to download a single dataset in specific format or all datasets at once.
Data format
PDB Data: structure derived from computed complex or chain in PDB format.
mmCIF Data: structure derived from computer complex or chain in mmCIF format.
CSV/JSON: Igzip compressed csv file with the following columns
1. Antibody-antigen complex
pdb_id, json, heavy, light, antigen
2. Heavy-light complex
pdb_id, json, heavy, light
3. Nanobody-antigen complex
pdb_id, json, nanobody, antigen
4. Protein-protein complex
pdb_id, json, protein1, protein1
5. Single chains (protein, heavy, light, nanobody)
pdb_id, json, chain, chain_type
Each row contains a json field with the schema below. Each entry has 'basic' attributes (Table 1) and structural attributes such as surface accessibility etc.
Dataset
"basic" field attributes
antibody_antigen_interface
pdb_id
heavy
heavy_seq
heavy_organism_name
heavy_taxonomy_id
heavy_host_organism_name
heavy_host_taxonomy_id
light
light_seq
light_organism_name
light_taxonomy_id
light_host_organism_name
light_host_taxonomy_id
antigen
antigen_seq
antigen_organism_name
antigen_taxonomy_id
antigen_host_organism_name
antigen_host_taxonomy_id
cl_id
l3_h3
method
resolution
last_update
initial_release
deposition_date
nanobody_antigen_interface
pdb_id
nanobody
nanobody_seq
nanobody_organism_name
nanobody_taxonomy_id
nanobody_host_organism_name
nanobody_host_taxonomy_id
antigen
antigen_seq
antigen_organism_name
antigen_taxonomy_id
antigen_host_organism_name
antigen_host_taxonomy_id
cl_id
l3_h3
method
resolution
last_update
initial_release
deposition_date
heavy_light_interface
pdb_id
heavy
heavy_seq
heavy_organism_name
heavy_taxonomy_id
heavy_host_organism_name
heavy_host_taxonomy_id
light
light_seq
light_organism_name
light_taxonomy_id
light_host_organism_name
light_host_taxonomy_id
cl_id
l3_h3
method
resolution
last_update
initial_release
deposition_date
protein_protein_interface
pdb_id
protein1
protein1_seq
protein1_organism_name
protein1_taxonomy_id
protein1_host_organism_name
protein1_host_taxonomy_id
protein2
protein2_seq
protein2_organism_name
protein2_taxonomy_id
protein2_host_organism_name
protein2_host_taxonomy_id
cl_id
l3_h3
method
resolution
last_update
initial_release
deposition_date
single_chain_*
pdb_id
chain
chain_seq
chain_organism_name
chain_taxonomy_id
chain_host_organism_name
chain_host_taxonomy_id
cl_id
l3_h3
method
resolution
last_update
initial_release
deposition_date
Delta/Parquet: for the purpose of using Apache Spark or other tools able to read parquet files the similar schema was used to dump datasets in the delta (delta.io) format.

Accessing the Database
We make the NAStructural DB free for non-commercial use by non-commercial entities. If you are a commercial entity and would like to employ the data in your activities, please get in touch with us.
Citing this work
We will post the associated manuscript when it clears reviews.
NAStructural DB: Structural database to facilitate computational studies of molecular modeling and recognition of proteins with special focus on antibody-antigen interactions.