Antibody Analytics
Accelerate your therapeutics pipeline with Antibody Analytics
Based on the comprehensive Antibody Knowledge Graph, we develop analytics solutions that connect the dots and provide insights into antibody developability, immunogenicity binding, and other features. Researchers use our solutions to quickly discover the advantages of individual candidate antibodies, stratify outputs of Next-Generation Sequencing (NGS) or phage display, and - most importantly - enhance their therapeutics pipelines thanks to data-driven insights into antibodies.Learn more
Sequence Engineering
Sequence Engineering module enables molecule developability optimization such as liability removal or humanization via combining two types of computational scores: Indicating possible issues with the molecule and Showing mutational choices to mitigate issues.
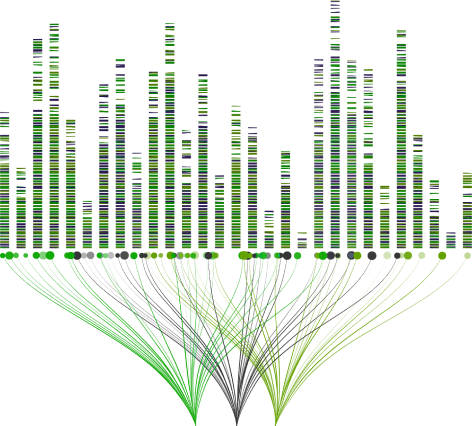
Batch Sequence Annotation with Clustering
Batch Sequence Annotation allows users to group sequences & annotate their liabilities. Users upload a set (batch) of antibody sequences in a well-adopted format (e.g., .fasta) and filter it by computational/machine learning attributes.
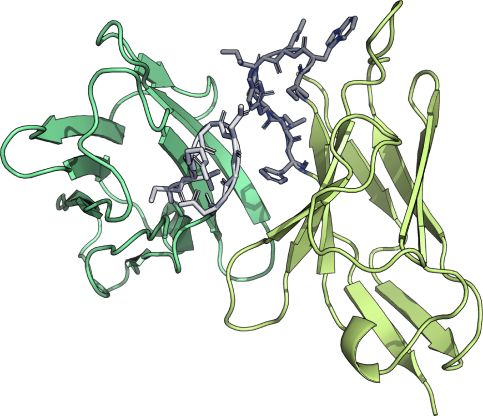
Structure Modeling
NaturalAntibody Structure Modeling allows therapeutic antibody lead optimization in the structural context. It offers information on what could be wrong with the molecule, where it could be modified, but offering a 3D context for the analysis.